1. Introduction
The generation of insulin-producing β-cells from human pluripotent stem cells is dependent on efficient endoderm induction and appropriate patterning and specification of this germ layer to a pancreatic fate. In this study, we elucidated the temporal requirements for TGFβ family members and canonical WNT signaling at these developmental stages and show that the duration of nodal/activin A signaling plays a pivotal role in establishing an appropriate definitive endoderm population for specification to the pancreatic lineage. WNT signaling was found to induce a posterior endoderm fate and at optimal concentrations enhanced the development of pancreatic lineage cells. Inhibition of the BMP signaling pathway at specific stages was essential for the generation of insulin-expressing cells and the extent of BMP inhibition required varied widely among the cell lines tested. Optimal stage-specific manipulation of these pathways resulted in a striking 250-fold increase in the levels of insulin expression and yielded populations containing up to 25% C-peptide+ cells.
2. Flow Chart
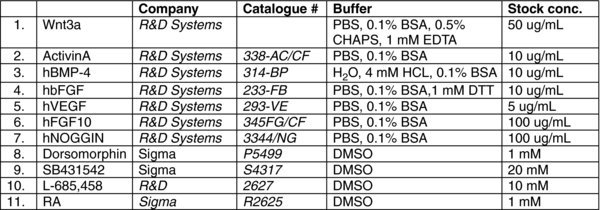
2.1. Reagent list
2.1.1. Reagents
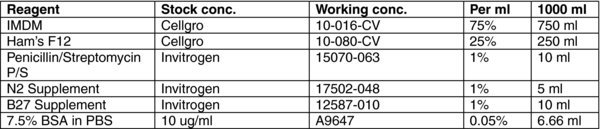
3. Reagent preparation
3.1. Stop medium

3.2. DNASE I (VWR, Cat # 80510-412, 10 MU)
Want final concentration to be 1 mg/ml
In the hood transfer powder to a 125 ml bottle
Bring the volume up to 153 ml with ice cold sterile water
Let dissolve on ice for 1–2 hours
Filter and aliquot 1 ml/eppendorf
Store at −20.
Filter sterilize, aliquot in 1 mL amounts and store frozen at −20°C
Use aliquots once and discard excess
3.3. L-ASCORBIC ACID (AA)(SIGMA # A-4544)
Prepare a stock solution of 5 mg/mL in cold TC-H2O. Leave on ice and vortex periodically until completely dissolved. Filter sterilize, aliquot and store at −20°C. Use once and discard
3.4. MONOTHIOGLYCEROL (MTG)(SIGMA# M-6145)
The amounts of MTG indicated in our protocols are recommended concentrations. However, it is important to test each new batch of MTG as there is variability between them. MTG should be aliquoted (1 mL) and stored frozen (−20°C). When aliquots are thawed, they can be used for several experiments and then discarded. Aliquoting of MTG is strongly recommended as it minimizes the amount of oxidation due to repeated opening.
3.5. TRANSFERRIN(ROCHE# 10 652 202)
The amounts of Transferrin indicated in our protocols are recommended concentrations. However, it is important to test each new batch of transferrin as there is variability between them. It should be aliquoted (2 mL) and stored at 4°C.
L-Glutamine (Gibco# 25030)
3.6. TRYPSIN-EDTA (Invitrogen# 25-053-CL)
3.7. Serum Free Differentiation (SFD) Media
RPMI (Gibco# 31800) supplemented with antibiotics, 10 mM HEPES and 1 mM Pyruvate.
3.8. (A) Day 0 Stage 1 MEDIUM
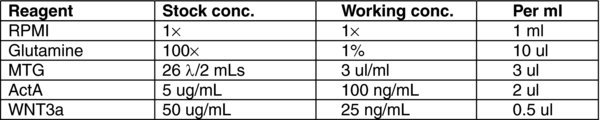
3.9. (B) Day 1–2 Stage 1 MEDIUM
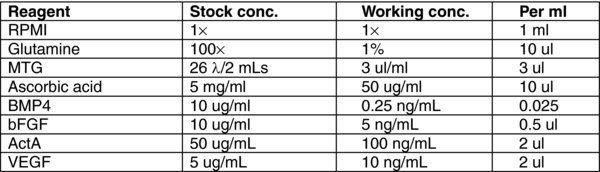
3.10. (C) Day 3, 5 Stage 2 MEDIUM
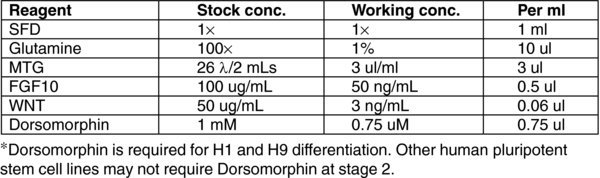
H21 (Gibco# 12800) with high D-glucose (4,500 mg/L).
3.11. (D) Day 6–8 Stage 3 MEDIUM
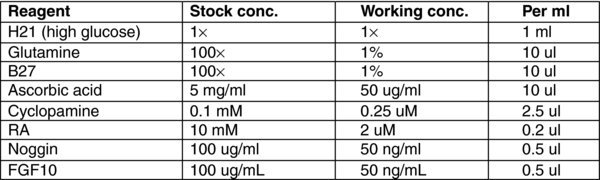
3.12. (E) Day 9,11 Stage 4 MEDIUM
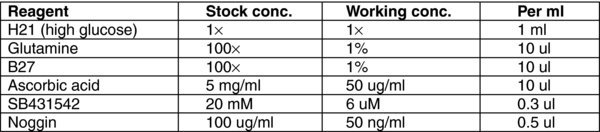
3.13. (F) Day 12–20 Stage 5 MEDIUM
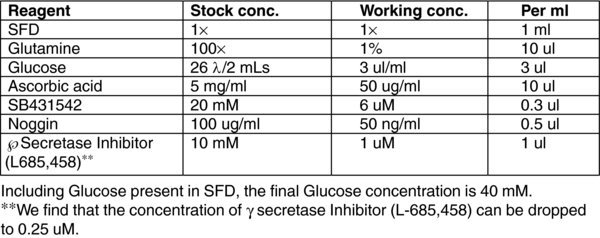
4. METHOD
4.1. Methods
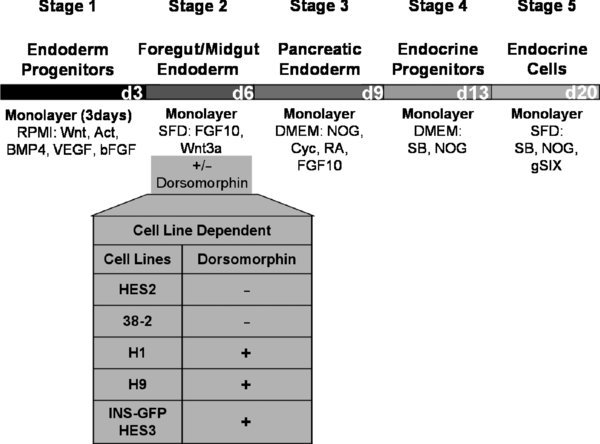
This protocol is designed for H1 and H9 human embryonic stem cell lines. Other lines may not require Dorsomorphin at stage 2 of differentiation.
4.2. hESC maintenance
Our lab routinely adapts hESCs to trypsin passage as this allows for easy passage and the maintenance and production of large numbers of cells. Successful maintenance of healthy undifferentiated hESCs is dependent on the appropriate concentrations of feeder cells and hESCs.
Mouse Embryonic Feeder Cells (MEFs). should be approximately 80% confluent and fresh, ideally cultured for only 24 hours prior to use. If the density of the MEFs is too high (confluent) the hESCs do not form discrete colonies but rather grow as disperse groups of cells, forming a monolayer. MEFs that are too sparce (<50%) do not provide adequate support for hESC maintenance. We routinely freeze irradiated MEFs at 2×106 cells per vial. Each vial contains enough cells for 18–24 wells of a twelve-well plate. The plating efficiency of each batch of MEFs needs to be tested.
hESCs. hESCs should be cultured at a density that allows the growth of distinct colonies with sharp borders within 4–5 days of culture. If the cells are too dense, the developing colonies grow into each other and form a monolayer. When too few cells are cultured, they can differentiate and tend to grow slower. Our stock of hESCs are frozen at 2×106 cells per vial. This concentration can be used for 6–24 wells of a twelve-well plate. The number of wells that can be cultured is dependent on the hESC line as well as the extent to which they are adapted to trypsin passage. Under optimal conditions with well adapted hESCs, you should be able to reach 70% confluency 4–5 days after plating, at this stage cells are ready to be differentiated.
Note: The protocol described below is designed to be carried out in a 12-well plate format.
4.3. Day 0: Stage 1 Endoderm Progenitors
Remove the medium from hESCs and wash once with RPMI.
To each well, add 1 mL of MEDIA (A). Incubate for 24 hours at 37°C in a 5% CO2 incubator.
4.4. Day 1–2: Stage 1 Endoderm Progenitors
There will be some debris in the cultures after 24 hours. Remove MEDIA A and wash once with RPMI.
To each well, add 1 mL of MEDIA (B). Incubate for 24 hours at 37°C in a 5% CO2 incubator.
Repeat steps 1–2 at day 2.
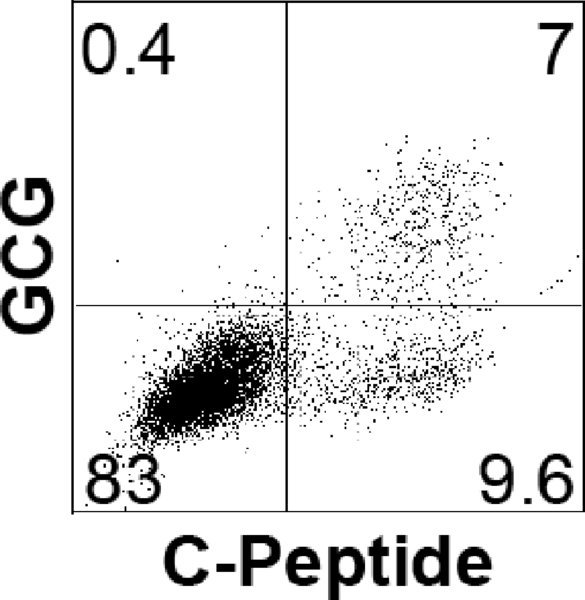
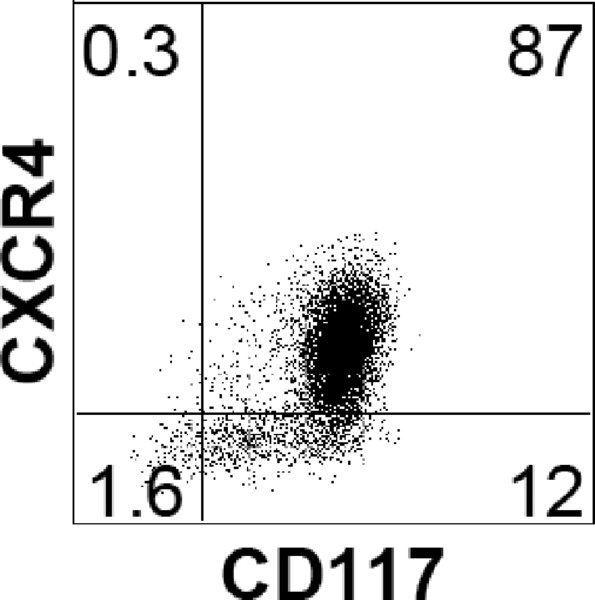
Note: Endoderm induction should be evaluated by flow cytometric analysis, monitoring the cells for expression of CXCR4 (CD184) and CD117 (c-KIT). As each hESC line has its own unique kinetics, it is best to define the endoderm stage based on the CXCR4/CD117 profile rather than by time in culture. The endoderm stage is defined by the appearance of a population that co-expresses CXCR4 and CD117. Using this protocol H1 gives rise to an average of 85% CXCR4+/CD117+ cells at day 3.
4.5. Day 3: Harvest for Flow Cytometry
Aspirate the medium and add 1 mL of TRYPSIN-EDTA. Incubate in a 37°C incubator for 2–3 minutes and then stop the reaction with 1 mL of STOP MEDIUM+DNase.
Spin for 5 min at 1000 RPM, aspirate and resuspend in PBS (−Ca2+ − Mg2+)+10%FCS (usually 500 uL per well harvested). Pass the cells through a 70 um filter to remove any clumps that are still remaining.
Stain with the desired antibodies (CXCR4, CD117) according to product datasheets and perform flow cytometric analysis.
4.6. Day 3, 5: Stage 2 Foregut/Midgut Endoderm
There will be some debris in the cultures after 24 hours. Remove MEDIA B and wash once with RPMI.
To each well, add 1 mL of MEDIA (C). Incubate for 48 hours at 37°C in a 5% CO2 incubator.
On day 5, remove MEDIA C.
To each well, add 1 mL of MEDIA (C). Incubate for 24 hours at 37°C in a 5% CO2 incubator.
4.7. Day 6–8: Stage 3 Pancreatic Endoderm
Remove Media C.
To each well, add 1 mL of MEDIA (D). Incubate for 24 hours at 37°C in a 5% CO2 incubator.
Repeat steps 1–2 on day 7 and 8.
Note: Due to the high instability of RA, we tend to feed in the dark and as fast as possible during stage 3.
4.8. Day 9,11: Stage 4 Endocrine Progenitors
Remove Media D.
To each well, add 1 mL of MEDIA (E). Incubate for 48 hours at 37°C in a 5% CO2 incubator.
On day 11, remove MEDIA E.
To each well, add 1 mL of MEDIA (E). Incubate for 48 hours at 37°C in a 5% CO2 incubator.
4.9. Day 13–20: Stage 5 Endocrine Cells
Remove Media E.
To each well, add 1 mL of MEDIA (F). Incubate for 72 hours at 37°C in a 5% CO2 incubator.
Feed every three days. During the course of this time hormone-expressing cells aggregate with each other and form clusters visible by eye.
Harvest at day 20.
Note: The percentage of endocrine cells should be evaluated by flow cytometric analysis, monitoring the cells for expression of C-Peptide and GCG. Below a typical profile for H1-differentiated cells at day 20.