Small non-coding RNAs bound to Argonaute (Ago) proteins have emerged as major players in epigenetic regulation of gene expression in plants and animals. The effector RNA molecules include the 21–25 nucleotide (ntd) long microRNAs (miRNAs) and the small interfering RNAs (siRNAs), which direct sequence specific target mRNA silencing. Based on phylogenetic analysis, Argonaute proteins can be subdivided into two subfamilies- Ago clade and Piwi clade. While, siRNAs and miRNAs associate with the Ago clade Argonautes, recent studies have identified a class of 24–30 ntd long Piwi-interacting RNAs (piRNAs), which bind to Piwi clade Argonautes. The piRNAs have important roles in germline development in flies, fish and mice. Here, we review recent studies bearing on the biological roles of piRNAs in the germline.
1. Introduction
Genetic analysis of developmental timing in C. elegans led to the discovery of the first small regulatory miRNA (Lee, Feinbaum et al., (1993)). We now know that miRNAs are involved in the spatial and temporal control of gene expression and control a myriad of physiological functions in both multicellular and unicellular organisms (Molnar, Schwach et al., (2007); Stefani and Slack (2008)). The miRNAs can be encoded by individual transcription units, polycistronic clusters, or the introns of pre-mRNAs (Bartel (2004)). Transcription by RNA Polymerase II produces long primary miRNAs (pri-miRNAs), double-stranded hairpin RNAs that are cleaved in the nucleus by an RNase III type enzyme, Drosha, to produce ∼65 ntd long pre-miRNAs (Lee, Jeon et al., (2002); Lee, Ahn et al., (2003)). Upon export into the cytoplasm by Exportin-5, pre-miRNAs are further processed by Dicer, another RNase III type endonuclease, to give ∼22 ntd mature miRNAs (Yi, Qin et al., (2003); Lund, Güttinger et al., (2004)). Endogenous siRNAs have now been described in plants, fungi, fission yeast, nematode worms, flies and mouse oocytes (Sontheimer and Carthew (2005); Vazquez (2006); Czech, Malone et al., (2008); Ghildiyal, Seitz et al., (2008); Kawamura, Saito et al., (2008); Okamura, Chung et al., (2008)). These 21–24ntd RNAs are produced from longer double-stranded RNAs (dsRNAs) by Dicer. In some cases, an RNA-dependent RNA polymerase (RdRp) –mediated step amplifies the original signal and leads to spreading of the signal beyond the trigger RNA sequence (Sijen, Fleenor et al., (2001)). Drosophila and mammals, however, lack RdRp enzymes. Both siRNAs and miRNAs bind to Argonaute proteins and downregulate target expression through sequence-specific cleavage or translational repression. In addition, both miRNAs and siRNAs are produced from dsRNA precursors by Dicer endonuclease mediated cleavage.
piRNAs, on the other hand, are longer (24–30 ntd) and do not require Dicer for their biogenesis (Vagin, Sigova et al., (2006); Houwing, Kamminga et al., (2007)). This newly described class of small RNAs derives its name from their effector proteins, which belong to the Piwi-clade of Argonautes. The Piwi clade is present in all the animals studied so far, and are most abundantly expressed in the germline (Carmell, Xuan et al., (2002)). The piRNAs were first identified through small RNA cloning experiments in Drosophila, where they are primarily derived from retrotransposons and other repetitive sequence elements and are required for retrotransposon silencing (Aravin, Lagos-Quintana et al., (2003)). As a result, these small RNAs were initially named repeat associated siRNAs (rasiRNAs). Subsequently, rasiRNAs were shown to bind to Piwi clade Argonautes, indicating that they are a class of piRNAs (Brennecke, Aravin et al., (2007); Yin and Lin (2007)).
piRNAs have now been cloned from flies, zebrafish and mammals, and they share some common features. They are often encoded in clusters, upto 100 kb in length. In Drosophila, piRNAs map to multiple regions in the genome, but in mammals, uniquely mapping piRNAs seem to be derived from large clusters which can account for most of the piRNAs. A striking feature of at least a subset of these clusters is a pronounced strand asymmetry, with essentially all of the piRNAs derived from either the plus or minus strand (Aravin, Gaidatzis et al., (2006); Vagin, Sigova et al., (2006); Houwing, Kamminga et al., (2007)). This finding suggests that piRNAs are derived from a long single-stranded precursor. However, piRNAs are derived from both strands in some clusters, and the nature of the precursor RNA remains to be defined. Deep sequencing of piRNA populations bound to their respective Piwi partners has shed some light on the mechanism of their biogenesis (Girard, Sachidanandam et al., (Girard, ; Sachidanandam, ., 2006); Brennecke, Aravin et al., (2007)). In flies, the Piwi proteins, Piwi and Aubergine (Aub) preferentially bind piRNAs that are antisense to the transposons, and they have a strong preference for a Uridine (U) at the 5’end. By contrast, Ago3 bound piRNAs are primarily derived from the sense strand of transposons. In addition, Ago3-piRNAs have an Adenine at the 10th nucleotide position, and are complementary to the first 10 bases of some Aub piRNAs. These findings suggest that plus strand piRNAs bound to Ago3 cleave minus strand precursor RNAs to generate the 5’ end of antisense piRNAs. A reciprocal reaction could generate the 5’ end of sense strand piRNAs, producing a “ping-pong” cleavage cycle of piRNA biogenesis (Brennecke, Aravin et al., (2007); Gunawardane, Saito et al., (2007)). However, there is no direct evidence for most of the steps in the cycle and the mechanism of piRNA production remains to be fully elucidated. Genetic studies have revealed a conserved function for Piwi-piRNA pathway in germline development (Cox, Chao et al., (2000); Carmell, Girard et al., (2007); Houwing, Kamminga et al., (2007); Klattenhoff, Bratu et al., (2007)). Below we review the role of piRNAs in the germline of flies, fish and mouse.
2. Germline development in Drosophila
The piRNA pathway has been studied extensively in Drosophila, where it is crucial to gametogenesis. In adult females, oogenesis initiates at the tip of the ovariole, a region called the germarium, where the germline stem cells (GSCs) divide asymmetrically to produce a daughter stem cell and a cystoblast. The cystoblast divides four times to produce 16 interconnected cells which form a single oocyte and 15 nurse cells. These 16 cell cysts are enveloped by somatic follicle cells and together they constitute an egg chamber, which buds from the germarium to join a linear arrangement of developing egg chambers. During stage 9 of oogenesis, asymmetric localization of RNA and protein morphogens begins, establishing the developmental axes. piRNA pathway mutations cause defects in both germline stem cell maintenance and embryonic axis specification (Cox, Chao et al., (2000); Cook, Koppetsch et al., (2004); Chen, Pane et al., (2007); Klattenhoff, Bratu et al., (2007)).
2.1. Germline stem cell maintenance in females
Mutations in the piRNA pathway gene piwi lead to germline stem cell loss (Fig. 1C) (Lin and Spradling (1997)). Clonal analyses indicate that Piwi is required in the somatic cells for germline stem cells maintenance, and in the germline for stem cell division (Cox, Chao et al., (1998); Cox, Chao et al., (2000)). It is unclear how Piwi expression in the soma ensures stem cell maintenance. However, Piwi protein associates with a subtelomeric region known as Telomeric-Associated Sequence (TAS) on the right arm of chromosome 3, which is the source of a subset of piRNAs. Transcription of this region is reduced in piwi mutants, and a P element transposon insertion near this region that increases expression of a 3R-TAS1 piRNA also suppresses stem cell loss in piwi mutants. These findings suggest that production of the 3R-TAS 1 piRNA is essential for germline stem cell maintenance (Yin and Lin (2007)). However, the suppressing P element insertion could have other effects on chromosome architecture or chromosome-chromosome interactions that are important for stem cell maintenance. In addition, it is unclear how expression of a piRNA suppresses loss of Piwi, which is presumably the effector protein in the piRNA complex. However, two additional Piwi clade Argonautes, Aubergine and Ago3, are expressed in the ovary, and these proteins could bind the 3R-TAS piRNA when Piwi is absent. Immunoprecipiation and deep sequencing experiments could be used to test this possibility. The function of Ago3 and Aubergine in germline stem cell division and maintenance has not been examined in detail.
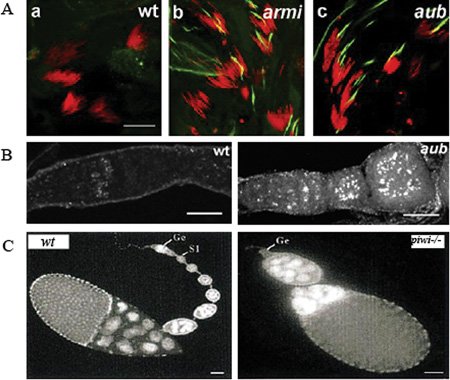
Figure 1. piRNA pathway mutants show germline defects in Drosophila . A.armi and aub mutant males show overexpression of Stellate protein in the testes. Testes were stained for DNA (red) and Stellate (green). Scale bar is 20 μM. Reprinted from Klattenhoff et al. (2007), with permission from Elsevier. B. DNA damage accumulation in the female germline of aub mutant germaria. Ovaries are labeled for γ-H2Av, a marker of DNA double-strand breaks. Scale bar is 20 μM. Reprinted from Klattenhoff et al. (2007), with permission from Elsevier. C.piwi is required for germline stem sell division during oogenesis. DAPi stained images of 0–1 day old, wt and piwi1 mutant ovaries showing loss of most of the developing egg chambers in piwi1, and germaria lacking germ lines (Ge). S1 refers to the first stage egg chamber budding off the germarium in the wt. Scale bar is 50 μM. Reprinted from Cox et al. (1998), with permission from Cold Spring Harbor Press Laboratory.
2.2. Oocyte axial patterning
Screens for mutations that disrupt embryonic axis specification identified aubergine, armitage, spindle-E, zucchini and squash, which disrupt posterior and dorsal-ventral patterning. All of these mutations were subsequently found to disrupt both piRNA production and retrotransposon silencing (Cook, Koppetsch et al., (2004); Chen, Pane et al., (2007); Klattenhoff, Bratu et al., (2007)). These findings raised the possibility that piRNAs control expression of genes in the axis specification pathway. However, mutations that disrupt meiotic DNA break repair trigger essentially identical axis specification defects (Ghabrial, Ray et al., (1998)), and the repair mutations disrupt axis specification through activation of DNA damage signaling via the ATR and Chk2 kinases (Ghabrial and Schupbach (1999)). These observations raised the possibility that piRNA mutations lead to ATR and Chk2 activation, rather than controlling axial patterning. Confirming this hypothesis, the axis specification defects in most piRNA mutations studied are dramatically suppressed by mutations in mei-41 and mnk, which encode ATR and Chk2, respectively (Chen, Pane et al., (2007); Klattenhoff, Bratu et al., (2007)). In addition, piRNA mutants accumulate γ-H2Av foci, which mark DNA double-strand breaks (Fig. 1B) (Klattenhoff, Bratu et al., (2007)). Mutations in mei-W68, which encodes the Spo11 nuclease that generates meiotic DNA breaks, do not suppress the formation of these foci or the embryonic patterning defects in armitage (Klattenhoff, Bratu et al., (2007)). The piRNA pathway thus does not appear to function in meiotic double strand break repair, but suppresses germline DNA damage by another mechanism.
piRNA pathway mutants lead to significant overexpression of retrotransposons in the female germline, suggesting that transposon mobilization could be the cause of DNA damage in the germline (Vagin, Klenov et al., (2004); Vagin, Sigova et al., (2006); Klenov, Lavrov et al., (2007)). Consistent with this hypothesis, transgenic expression of LINE retrotransposons leads to DNA damage in mammalian cells (Belgnaoui, Gosden et al., (2006); Gasior, Wakeman et al., (2006)). However, DNA damage can also lead to loss of transposon silencing (Farkash, Kao et al., (2006)). It is therefore possible that piRNA mutations lead to DNA damage by a transposon independent mechanism, and that the resulting damage triggers the observed loss of retrotransposon silencing. Molecular characterization of the DNA breaks in piRNA pathway mutants would help resolve this issue.
2.3. Hybrid dysgenesis and trans-silencing
Hybrid dysgenesis is a genetic phenomenon observed in crosses between Drosophila males carrying specific transposons and naïve females that lack these transposons. It is characterized by reduced fertility, transposon mobilization and high mutation rates in the progeny (Kidwell, Kidwell et al., (1977); Engels and Preston (1979)). Transposon expression is silenced in the few surviving progeny, and the ability to silence is maternally inherited. Studies in Drosophila virilis suggest that this silencing is mediated by 21–25 ntd small RNAs (Blumenstiel and Hartl (2005)). Another set of studies on hybrid crosses suggest that the ability to repress the P element is dependent on aubergine, and the heterochromatin protein 1 (HP1) (Reiss, Josse et al., (2004)). The telomeric trans-silencing effect (TSE) is a related process in which a transgene in the euchromatin is silenced by a homologous transgene inserted into subtelomeric heterochromatin (Reiss, Josse et al., (2004); Josse, Teysset et al., (2007)). This process is disrupted by piRNA pathway mutations, suggesting that piRNAs are generated from transposons inserted into the sub-telomeric heterochromatin, and that the resulting Piwi-piRNA complexes silence homologous transposons at other chromosome localizations. However, piRNAs corresponding to the telomeric transposon have not yet been identified, and it is unclear if euchromatic transposon silencing is at the transcriptional, co-transcriptional, or post-transcriptional level.
2.4. Germline determination in the early embryo
Aub and Piwi also localize to the posterior pole of the oocyte and appear to associate with polar granules, which are required for germline specification and are incorporated into the germline precursor cells (pole cells) during early embryogenesis (Harris and Macdonald (2001); Megosh, Cox et al., (2006)). Maternally inherited Aub and Piwi could therefore maintain piRNA based silencing during early germline development. Piwi, in complex with Dicer-1 and Fragile-X Mental Retardation Protein (FMRP), known miRNA components, also is a part of the pole plasm and is involved in germline determination (Megosh, Cox et al., (2006)).
2.5. Additional piRNA functions in Drosophila males
Mutations in piRNA pathway genes lead to a loss of Stellate locus silencing during spermatogenesis (Vagin, Klenov et al., (2004); Vagin, Sigova et al., (2006)). The Stellate protein is encoded by repeats on the X chromosome, and Stellate expression is suppressed in a homology-dependent manner by the Y-linked locus-Suppressor of Stellate (Su(Ste)). piRNAs homologous to the Stellate locus are produced from the Su(Ste) region, and Su(Ste) mutations lead to overexpression of Stellate protein, which forms crystals in the testis (Fig. 1A) (Vagin, Klenov et al., (2004)). Mutations in piwi also disrupt retrotransposon silencing and lead to mobilization of mdg1 retrotransposons in the male germline (Kalmykova, Klenov et al., (2005)). Some piRNA pathway mutations lead to partial loss of male fertility (N. Schultz and W. Theurkauf, unpublished observations), but whether these defects are related to Sellate over-expression, transposon mobilization, or changes in the expression of unidentified piRNA targets, is currently unknown.
3. Germline development in Zebrafish
3.1. Germ cell maintenance in Zebrafish
The zebrafish genome encodes two Piwi homologs, Ziwi and Zili. Based on sequence similarity, Ziwi appears to be the ortholog of mouse MIWI, and Zili is the ortholog of MILI (Houwing, Kamminga et al., (2007)). Ziwi localizes to the perinuclear nuage, which appears to represent germ granules and specifies the primordial germ cells during early embryogenesis (Tan, Lee et al., (2002)). piRNAs are found in both testes and ovaries, and their localization and temporal pattern of expression are coincident with Ziwi. A significant fraction of these piRNAs derive from transposable elements, suggesting that this pathway also silences transposons in zebrafish. Ziwi-bound piRNAs have a similar 5′ end modification and length as their cousins in Drosophila and mammals (Houwing, Kamminga et al., (2007)). As observed in mouse Piwi mutants, ziwi mutants show a progressive decline in the germ cells due to apoptosis (Fig. 2A,B). Another striking feature of loss of Ziwi is that the mutants are all phenotypically males. This defect, however, seems to be a secondary consequence of loss of primordial germ cells (PGCs) which are required during early embryogenesis for female development (Slanchev, Stebler et al., (2005)).
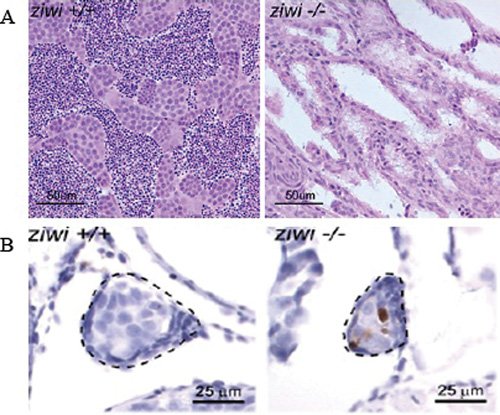
Figure 2. piRNA pathway is required for germ-cell maintenance in Zebrafish. A. Germ cell loss in ziwi-/- testis. Hematoxylin and eosin staining of wild-type (left) and ziwi-/- (right) testis, showing germ-cell depleted testis in ziwi-/-.B. Increased apoptosis, as witnessed by Caspase-3 staining (brown) in the developing germline (indicated) of the ziwi-/- gonads, at 21 days of development. Panels A and B reprinted from Houwing et al. (2007), with permission from Elsevier.
4. Germline development in mouse
4.1. Mouse spermatogenesis
The mouse genome encodes three Piwi homologs, MIWI (PIWIL1), MILI (PIWIL2) and MIWI2 (PIWIL4). All three of these proteins have distinct spatial and temporal expression patterns in the male germline. Knockout animals in any of the mouse piwi genes disrupt male germline development (Deng and Lin (2002); Kuramochi-Miyagawa, Kimura et al., (2004); Carmell, Girard et al., (2007)). Mouse spermatogenesis initiates at day 3 after birth (de Rooij and Grootegoed (1998)). The primordial germ cells, after reaching the gonad, form spermatogonia, subsets of which are stem cells capable of self-renewal. The stem cells then differentiate to produce primary spermatocytes up to day 6. At day 10, the primary spermatocytes enter meiosis I and yield a pair of secondary spermatocytes, which complete the second meiotic division. The haploid cells from the meiotic process are called spermatids, which undergo the process of sperm maturation or spermiogenesis, to yield mature sperm. While Mili and Miwi2 null mice show meiotic arrest, Miwi null mice show defects in spermiogenesis (Fig. 3B) (Deng and Lin (2002); Kuramochi-Miyagawa, Kimura et al., (2004); Carmell, Girard et al., (2007)). These knockout phenotypes correlate well with the expression pattern of the Piwi proteins. Mutation in any of the piwi genes causes loss of germ cells due to increased apoptosis (Fig. 3A). Miwi2 mutant spermatocytes show an increased accumulation of γ-H2AX during zygotene, which indicates the presence of unrepaired meiotic double-strand breaks and/or DNA damage due to other reasons (Fig. 3C) (Carmell, Girard et al., (2007)). For example, increased γ-H2AX staining has been observed in mutants defective in synapsis. A peculiar feature of mammalian male meiosis is the formation of a dense sex body by the XY bivalent that undergoes transcriptional silencing. The sex body is positive for γ-H2AX and other DNA repair proteins, including BRCA1, RAD51 and RPA. However, Miwi2 mice fail to stain for γ-H2AX during pachytene, suggesting that sex body formation is disrupted. Interestingly, some piRNAs or their precursors have been localized to regions on the sex body (Marcon, Babak et al., (2008)). These findings sugget that the piRNA pathway may be involved in the transcriptional silencing of the XY bivalent.
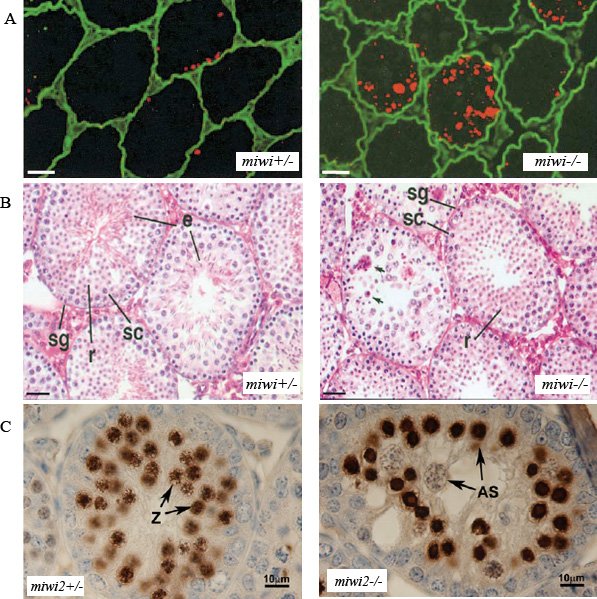
Figure 3. piRNA pathway is required for spermatogenesis in mouse. A. High levels of apoptosis in miwi-/- testes, as judged by the TUNEL (red) labeling. Scale bar is 25μM. B. Spermatogenesis arrest in miwi-/- at the early round spermatid stage. Hematoxylin/Eosin staining of miwi+/- and miwi-/- testes show the absence of elongated spermatid (e) in miwi-/- and the presence of fragmented nuclei (arrows). Stages of spermatogenesis labeled here- spermatogonia (sg), spermatocytes (sc), round spermatid (r). Scale bar is 25 μM. Panels A and B reprinted from Deng and Lin ((2002)) with permission from Elsevier. C. Increased DNA damage accumulation in miwi2-/- testis. Intense staining of γ-H2AX (brown) and presence of abnormal spermatocytes (AS) in seminiferous tubules from 14 day old miwi2-/- animals. Zygotene cells (z) are marked in the miwi2+/- and absent in the miwi2-/- null mice. Reprinted from Carmell et al. (2007), with permission from Elsevier.
Although mammalian piRNAs are depleted of repetitive sequences, a detailed analysis of developmentally expressed MILI-bound piRNAs revealed two distinct populations and a significant number of piRNA clusters correspond to repeats (Aravin, Sachidanandam et al., (2007)). In addition, some transposons are demethylated in the Mili and Miwi2 mice, suggesting that they are transcriptionally active (Aravin, Sachidanandam et al., (2007); Carmell, Girard et al., (2007)). These findings suggest that the mammalian Piwi-piRNA pathway, like the Drosophila piRNAs, may be required for transposon silencing.
MIWI and its piRNAs are found in the polysome fractions (Grivna, Pyhtila et al., (2006)). Additionally, MIWI is found in ribonuclear protein fractions along with a testis-expressed kinesin, KIF17b, and binds the mRNAs of ACT and CREM target genes, which are the master regulators of spermiogenesis (Kotaja, Lin et al., (2006)). Consistent with this, Drosophila Aub has been implicated in translational regulation of some targets like Oskar. The presence of MIWI piRNAs against the target mRNAs would give a much more direct evidence for the role of this pathway in translational control.
In contrast to flies and zebrafish, the mouse piRNA pathway mutations do not disrupt the female germline. This is because retrotransposon silencing in the mouse oocytes seems to be largely dependent on the endogenous RNAi pathway. Endogenous siRNAs, with sequence homology to retroelements, have been cloned from growing mouse oocytes. Consistent with this observation is the disruption of transposon silencing in the conditional Dicer or Ago2 mutants. Therefore, piRNAs primarily silence transposons in the male germline, and a distinct class of endogenous siRNAs silence transposons during oogenesis (Watanabe, Takeda et al., (2006)).
5. Summary and perspective
In every system studied to date, Piwi clade Argonautes, and presumably piRNAs, are required for germline development. Studies in Drosophila, zebrafish and mouse link these defects to loss of transposon silencing, and the majority of Drosophila piRNAs are derived from transposons and other repeated elements. These findings suggest that the piRNA pathway prevents mobilization of selfish genetic elements, which can trigger DNA damage and cell death. However, DNA damage can induce transposon mobilization, and the nature of the DNA lesions that accumulate in piRNA mutants has not been defined. piRNAs may therefore have a primary function in genome maintenance that is independent of transposon silencing, which could be secondary to DNA break formation. The mechanism of piRNA mediated silencing also remains to be established. There are data supporting both transcriptional and post-transcriptional mechanisms, and it is possible that these RNAs function in both the nucleus and cytoplasm.
The mechanism of piRNA biogenesis also remains to be elucidated. The sequence of piRNAs bound to Piwi class Argonautes suggest a ping-pong model for biogenesis, based on Piwi protein mediated cleavage. However, the vast majority of enzymes in this hypothetical pathway have not been identified and precursor RNAs have not been detected. Furthermore, the relationship between piRNA abundance and function remains enigmatic. In Drosophila, the most abundant piRNAs are derived from the roo retrotransposons, but roo silencing is only minimally disrupted by piRNA pathway mutations. By contrast, HeT-A piRNAs are relatively rare, but expression of this element increases several hundred fold when piRNA production is disrupted. In mammals, the vast majority of piRNAs are derived from intergenic regions with no known function. Thus, it seems that the piRNA pathway in mammals has evolved beyond its function as a transposon mobilization checkpoint. Elucidation of the functional targets for the most abundant piRNAs would shed some light on their additional roles. Available genetic data demonstrate a conserved function for piRNAs in germline development. However, the studies to date raise as many questions as answers about the function and biogenesis of this new class of small RNAs.
Copyright: © 2008 Jaspreet S. Khurana and William E. Theurkauf.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
* Correspondence should be addressed to: Jaspreet S. Khurana
* Edited by Patricia Donahoe and Haifan Lin. Last revised June 6, 2008. Published July 30, 2008. This chapter should be cited as: Khurana, J.S. and Theurkauf, W.E., piRNA function in germline development (July 30, 2008), StemBook, ed. The Stem Cell Research Community, StemBook, doi/10.3824/stembook.1.12.1, https://www.stembook.org .
References
- Aravin, A. A. Lagos-Quintana, M. (2003). The small RNA profile during Drosophila melanogaster development. Dev Cell 5(2), 337–50.
- Aravin, A. Gaidatzis, D. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature advanced online publication (7099), 203–207.
- Aravin, A. A. Sachidanandam, R. (2007). Developmentally regulated piRNA clusters implicate MILI in transposon control. Science (New York, N.Y.) 316(5825), 744–747.
- Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2), 281–97.
- Belgnaoui, S. M. Gosden, R. G. (2006). Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer cell international 6, 13.
- Blumenstiel, J. P. Hartl, D. L. (2005). Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proceedings of the National Academy of Sciences of the United States of America 102(44), 15965–15970.
- Brennecke, J. Aravin, A. A. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128(6), 1089–1103.
- Carmell, M. A. Xuan, Z. (2002). The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & development 16(21), 2733–2742.
- Carmell, M. A. Girard, A. (2007). MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Developmental cell 12(4), 503–514.
- Chen, Y. Pane, A. (2007). Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Current biology: CB 17(7), 637–642.
- Cook, H. Koppetsch, B. (2004). The Drosophila SDE3 Homolog armitage Is Required for oskar mRNA Silencing and Embryonic Axis Specification. Cell 116(6), 817–829.
- Cox, D. N. Chao, A. (1998). A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12(23), 3715–27.
- Cox, D. N. Chao, A. (2000). piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127(3), 503–14.
- Czech, B. Malone, C. (2008). An endogenous small interfering RNA pathway in Drosophila. Nature.
- Deng, W. Lin, H. (2002). miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2(6), 819–30.
- Engels, W. R. Preston, C. R. (1979). Hybrid dysgenesis in Drosophila melanogaster: the biology of female and male sterility. Genetics 92(1), 161–174.
- Farkash, E. A. Kao, G. D. (2006). Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic acids research 34(4), 1196–1204.
- Gasior, S. L. Wakeman, T. P. (2006). The human LINE-1 retrotransposon creates DNA double-strand breaks. Journal of molecular biology 357(5), 1383–1393.
- Ghabrial, A. Ray, R. P. (1998). okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev 12(17), 2711–23.
- Ghabrial, A. Schupbach, T. (1999). Activation of a meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nat Cell Biol 1(6), 354–7. [java/Propub/cellbio/ncb1099_354.fulltext java/Propub/cellbio/ncb1099_354.abstract]
- Ghildiyal, M. Seitz, H. (2008). Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science (New York, N.Y.) 320(5879), 1157396–1081.
- Girard, ; Sachidanandam, . (2006). A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature advanced online publication (7099), 199–202.
- Grivna, S. T. Pyhtila, B. (2006). MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proceedings of the National Academy of Sciences of the United States of America 103(36), 13415–13420.
- Gunawardane, L. S. Saito, K. (2007). A slicer-mediated mechanism for repeat-associated siRNA 5’ end formation in Drosophila. Science (New York, N.Y.) 315(5818), 1587–1590.
- Harris, A. N. Macdonald, P. M. (2001). Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 128(14), 2823–32.
- Houwing, S. Kamminga, L. M. (2007). A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129(1), 69–82.
- Josse, T. Teysset, L. (2007). Telomeric trans-silencing: an epigenetic repression combining RNA silencing and heterochromatin formation. PLoS Genetics 3(9), 1633–1643.
- Kalmykova, A. I. Klenov, M. S. (2005). Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic acids research 33(6), 2052–2059.
- Kawamura, Y. Saito, K. (2008). Drosophila endogenous small RNAs bind to Argonaute[thinsp]2 in somatic cells. Nature.
- Kidwell, M. G. Kidwell, J. F. (1977). Hybrid Dysgenesis in DROSOPHILA MELANOGASTER: A Syndrome of Aberrant Traits Including Mutation, Sterility and Male Recombination. Genetics 86(4), 813–833.
- Klattenhoff, C. Bratu, D. P. (2007). Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Developmental cell 12(1), 45–55.
- Klenov, M. S. Lavrov, S. A. (2007). Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic acids research 35(16), 5430–5438.
- Kotaja, N. Lin, H. (2006). Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. Journal of cell science 119(Pt 13), 2819–2825.
- Kuramochi-Miyagawa, S. Kimura, T. (2004). Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development (Cambridge, England) 131(4), 839–849.
- Lee, R. C. Feinbaum, R. L. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5), 843–54.
- Lee, Y. Jeon, K. (2002). MicroRNA maturation: stepwise processing and subcellular localization. Embo J 21(17), 4663–70.
- Lee, Y. Ahn, C. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature 425(6956), 415–9.
- Lin, H. Spradling, A. C. (1997). A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124(12), 2463–76.
- Lund, E. Güttinger, S. (2004). Nuclear export of microRNA precursors. Science (New York, N.Y.) 303(5654), 95–98.
- Marcon, E. Babak, T. (2008). miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology 16(2), 243–260.
- Megosh, H. B. Cox, D. N. (2006). The role of PIWI and the miRNA machinery in Drosophila germline determination. Current biology: CB 16(19), 1884–1894.
- Molnar, A. Schwach, F. (2007). miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature advanced online publication (7148), 1126–1129.
- Okamura, K. Chung, W.-J. (2008). The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature.
- Reiss, D. Josse, T. (2004). aubergine mutations in Drosophila melanogaster impair P cytotype determination by telomeric P elements inserted in heterochromatin. Molecular genetics and genomics : MGG 272(3), 336–343.
- Sijen, T. Fleenor, J. (2001). On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107(4), 465–76.
- Slanchev, K. Stebler, J. (2005). Development without germ cells: the role of the germ line in zebrafish sex differentiation. Proceedings of the National Academy of Sciences of the United States of America 102(11), 4074–4079.
- Sontheimer, E. J. Carthew, R. W. (2005). Silence from within: endogenous siRNAs and miRNAs. Cell 122(1), 9–12.
- Stefani, G. Slack, F. (2008). Small non-coding RNAs in animal development. Nature reviews. Molecular cell biology 9(3), 219–230.
- Tan, C.-H. Lee, T.-C. (2002). Ziwi, the zebrafish homologue of the Drosophila piwi: co-localization with vasa at the embryonic genital ridge and gonad-specific expression in the adults. Mechanisms of development 119(Suppl 1), S221–4.
- Vagin, V. V. Klenov, M. S. (2004). The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA biology 1(1), 54–58.
- Vagin, V. V. Sigova, A. (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313(5785), 320–324.
- Vazquez, F. (2006). Arabidopsis endogenous small RNAs: highways and byways. Trends in plant science 11(9), 460–468.
- Watanabe, T. Takeda, A. (2006). Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes & development 20(13), 1732–1743.
- Yi, R. Qin, Y. (2003). Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & development 17(24), 3011–3016.
- Yin, H. Lin, H. (2007). An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature advanced online publication (7167), 304–308.
- de Rooij, D. G. Grootegoed, J. A. (1998). Spermatogonial stem cells. Current opinion in cell biology 10(6), 694–701.

