Pluripotent stem cells, such as Embryonic Stem cells, have the extraordinary ability to give rise to all cell types of the body, and hold great potential for regenerative medicine. Pluripotent stem cells are derived from in vitro culture of early embryos or by over-expression of specific transcription factors in somatic cells. In recent years great progress has been made towards understanding the molecular basis of pluripotency. The transcriptomes of various pluripotent stem cell lines have been examined in order to identify molecular mechanisms that control self-renewal and pluripotency. These studies have begun to reveal the components and architecture of the transcriptional regulatory networks of pluripotency, and suggest that transcription factors maintain the pluripotent state in the backdrop of a loose, accessible chromatin. Such a chromatin state may allow for increased global transcription, resulting in the expression of many novel and as yet uncharacterized transcripts. Fluctuations in the transcriptional networks may alternate pluripotent stem cells between states with distinct probabilities for self-renewal or differentiation. In this chapter, we review studies of the transcriptome of pluripotent stem cells, highlight intriguing transcriptional similarities between pluripotent stem cells, cancer cells and the germline, and discuss future avenues of research.
1. Introduction
Embryonic Stem (ES) cells are the prototypical pluripotent stem cell: they are able to both self-renew and differentiate into all cell types. Because of these properties, ES cells are an excellent system to study cellular differentiation in both normal and diseased states. In addition, cells derived from ES cells have great potential for therapeutic applications, provided they can be efficiently expanded and differentiated in vitro. An important step towards therapeutic applications of ES cells is to identify the molecules and pathways governing ES cell proliferation and differentiation.
ES cells are derived from in vitro culture of the inner cell mass (ICM) of the blastocyst, prior to implantation. Another pluripotent cell type, Embryonic Germ (EG) cells, can be derived from in vitro culture of the embryonic germline, Primordial Germ Cells (PGCs). Recently, induced Pluripotent Stem (iPS) cells have been derived from non-embryonic somatic cells by the introduction of specific key transcription factors (Aoi et al., 2008; Blelloch et al., 2007; Hanna et al., 2008; Lowry et al., 2008; Maherali et al., 2007; Meissner et al., 2007; Okita et al., 2007; Park et al., 2008; Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Wernig et al., 2007; Yu et al., 2007). Both embryo-derived (ES and EG) and non-embryo-derived (iPS) pluripotent cells share highly similar transcriptional profiles (Sharova et al., 2007; Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Wernig et al., 2007; Yu et al., 2007).
In recent years there has been great progress towards understanding the molecular basis of pluripotency. The transcriptomes of various pluripotent cell lines have been extensively examined in order to identify molecular factors and mechanisms that control self-renewal and differentiation. Genome-wide expression profiling using expressed sequence tag (EST) analysis (Brandenberger et al., 2004), serial analysis of gene expression (SAGE; Richards et al., 2004), massively parallel signature sequencing (MPSS; Brandenberger et al., 2004; Wei et al., 2005) and DNA microarrays (Abeyta et al., 2004; Bhattacharya et al., 2004; Ivanova et al., 2002; Ramalho-Santos et al., 2002; Sato et al., 2003; Sperger et al., 2003; see Figure 1) have uncovered gene networks and signaling pathways believed to be essential for maintenance of the pluripotent state. Here, we review studies of the transcriptome of pluripotent stem cells, and highlight intriguing similarities between pluripotent stem cells, cancer cells and the germline.
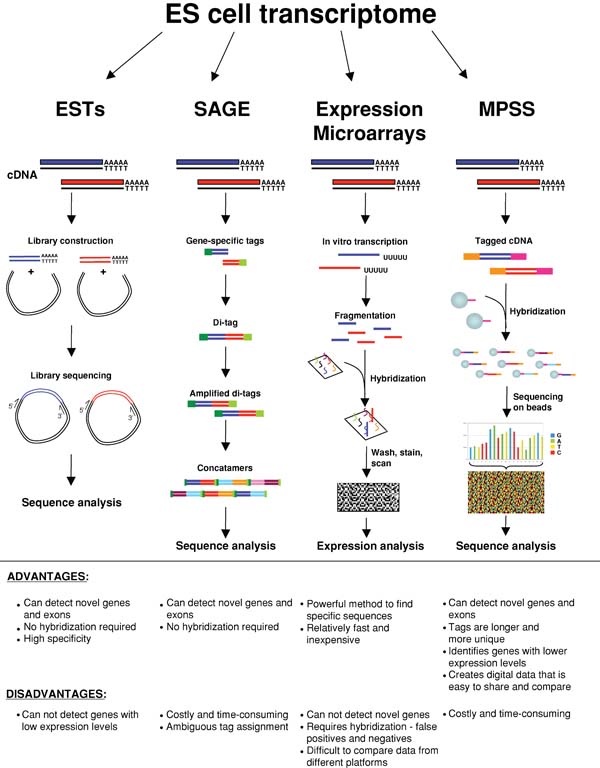
Genome-wide expression profiling using expressed sequence tag (EST) analysis, serial analysis of gene expression (SAGE), DNA microarrays and massively parallel signature sequencing (MPSS) have all been used to study the transcriptome of ES cells. ESTs are generated by inserting cDNA into cloning vectors to produce a cDNA library. The inserts are sequenced and the resulting EST sequences are analyzed. SAGE involves the generation of gene-specific tags, typically 10–14 basepairs in length. These tags are ligated to form di-tags, amplified by PCR, ligated again to form concatamers and sequenced. Expression microarrays are arrays of synthesized oligonucleotides or spotted cDNAs in a predetermined spatial orientation. Total RNA is reverse transcribed, fluorescently labeled and hybridized to the microarray. Specific hybridization signals are detected by a fluorescent scanner, and gene expression is deduced. Tiling arrays are modified microarrays where all non-repetitive genomic DNA is represented at various sequence resolutions. Unlike conventional microarrays, tiling arrays enable the discovery of novel transcribed sequences and regulatory elements through the unbiased examination of the genome. MPSS involves the cloning of a cDNA library on beads and the acquisition of 17–20-nucleotide tags from these cDNAs using an unconventional sequencing method that allows for large-scale sequencing. Recently developed deep sequencing approaches using 454 and Solexa platforms promise to provide a comprehensive description of the pluripotent transcriptome.
2. Molecular signature of pluripotent stem cells
To define the molecular signature of pluripotent stem cells, several groups compared the expression profiles of ES cells to those of differentiated cells (Bhattacharya et al., 2004; Brandenberger et al., 2004; Liu et al., 2006; Miura et al., 2004; Zhou et al., 2007), to other stages of the developing embryo (Falco et al., 2007; Sharov et al., 2003), to other types of pluripotent and adult stem cells (Abeyta et al., 2004; Ivanova et al., 2002; Ramalho-Santos et al., 2002; Sharova et al., 2007; Zeng et al., 2004), or to ES cells of other species (Sato et al., 2003; Sun et al., 2007; Wei et al., 2005). An emerging picture from these studies is that some genes are predominantly or exclusively expressed in pluripotent cells, such as Oct4, Nanog, Rex1 and Utf1. These studies also point to the activity of particular signaling pathways in ES cells, such as Jak/Stat, Bmp, Wnt and FGF. Many of these studies highlight an over-representation of transcription, chromatin remodeling and DNA repair factors in the pluripotent transcriptome, suggesting that these play an essential role in pluripotency. Upon in vitro differentiation of ES cells, pluripotency-associated genes are consistently down-regulated regardless of the method of differentiation, while the induced genes depend on the method of differentiation (Walker et al., 2007).
Predominant or exclusive expression of a gene in ES cells provides insufficient evidence for a role of that gene in self-renewal or pluripotency. Some genes predominantly expressed in ES cells, such as Oct4 or Sox2, had been shown to be essential regulators of pluripotency before transcriptional profiling became widely used (Avilion et al., 2003; Nichols et al., 1998). By combining expression profiling with RNA interference (RNAi), Ivanova and colleagues identified several novel regulators of ES cell self-renewal, such as Esrrb, Tbx3 and Tcl1 (Ivanova et al., 2006). In addition, gene expression profiling of ES cells in which expression of a critical transcription factor has been perturbed can provide insight into the transcription regulatory networks of pluripotency (Ivanova et al., 2006; Matoba et al., 2006; Walker et al., 2007). For example, genes that are affected by downregulation of Oct4 are similarly affected by downregulation of Nanog or Sox2, indicating that Oct4, Sox2 and Nanog cooperate to control transcription of target genes (Ivanova et al., 2006; Walker et al., 2007). Downregulation of Esrrb, Tbx3 and Tcl1 affected the expression of a different set of genes, suggesting that they regulate a distinct ES cell pathway (Ivanova et al., 2006). Thus, transcriptional profiling, in particular in the setting of gene-manipulated ES cells, will likely continue to provide important insights into the regulation of the pluripotent transcriptome.
3. Co-expression to co-regulation: regulatory pathways
Genes co-expressed in pluripotent cells are likely to be (at least in part) co-regulated by the same transcription factors. Several key transcription factors have been identified as expressed in ES cells and involved in the maintenance of pluripotency. Among these are factors known as “core regulators of pluripotency”: Oct4, Sox2 and Nanog (see Figure 2).
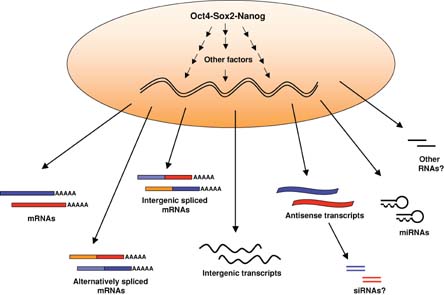
The “core” regulators of pluripotency Oct4, Sox2 and Nanog, act in concert with other transcription and chromatin modifying factors to establish and maintain the transcriptional program of pluripotency. The accessible chromatin of ES cells may allow for increased global transcription resulting in expression of many genes, their splicing isoforms, intergenic spliced mRNAs, non-coding intergenic transcripts and antisense transcripts. The function of these novel transcripts in ES cells is not yet understood. Antisense transcripts are thought to regulate the expression of their sense counterparts, possibly by sense-anti-sense pairing and production of siRNAs. Endogenous siRNAs have been recently detected in oocytes (Tam et al., 2008; Watanabe et al., 2008), but so far they have not been described in ES cells. miRNAs however, have been shown to be expressed in ES cells and are required for their proliferation and differentiation (reviewed in (Bibikova et al., 2008)).
The POU-family transcription factor Oct4 is predominantly expressed in pluripotent cells of the developing mouse embryo (ICM, PGCs) and their in vitro counterparts (ES and EG cells, respectively; Pesce and Scholer, 2001). Embryos lacking Oct4 die at the blastocyst stage, and ES cells with perturbed levels of Oct4 expression exit self-renewal and differentiate (Nichols et al., 1998; Niwa et al., 2000). The Sry-related transcription factor Sox2 can act synergistically with Oct4 to activate Oct-Sox enhancers, which regulate the expression of pluripotent stem cell-specific genes, including Nanog, Oct4 and Sox2 itself (Chew et al., 2005; Kuroda et al., 2005; Nishimoto et al., 2005; Okumura-Nakanishi et al., 2005; Rodda et al., 2005; Tomioka et al., 2002). A recent report indicates that the essential function of Sox2 may be to stabilize ES cells in a pluripotent state by maintaining the appropriate level of Oct4 expression (Masui et al., 2007).
Similarly to Oct4, the homeobox transcription factor Nanog is predominantly expressed in ICM, PGCs, ES and EG cells. Nanog has been shown to sustain pluripotency in ICM and ES cells, and to support self-renewal of mouse ES cells in the absence of LIF, a cytokine essential for mouse ES cells (Chambers et al., 2003; Mitsui et al., 2003). Recent genome-wide studies using chromatin immunoprecipitation combined with microarrays (ChIP-chip) or paired-end ditag sequencing (ChIP-PET) revealed Nanog as part of a transcriptional network that regulates the activity of numerous genes in collaboration with Oct4 and Sox2 (Boyer et al., 2005; Loh et al., 2006). Since each of these three genes contains binding sites for each of the three transcription factors, it was suggested that they may act as a self-organizing feed-forward loop that sustains the expression levels needed to maintain pluripotency. However, Chambers et al recently showed that it is possible to derive ES cells that lack Nanog but still retain the expression of pluripotency markers, including Oct4 and Sox2 (Chambers et al., 2007). Although they are prone to differentiate, these ES cells are capable of self-renewal in the absence of Nanog, and they can contribute to all three germ layers in chimeras. Therefore, it appears that, rather than being essential for establishing pluripotency, Nanog may function to reinforce the stability of the pluripotent state.
To delineate regulatory relationships between Oct4, Sox2 and Nanog and their targets, Walker and colleagues combined available ChIP-chip data (Boyer et al., 2005; Boyer et al., 2006; Loh et al., 2006) with time-course gene expression profiling of differentiating ES cells, as well as ES cells deficient for Oct4 or Sox2 (Walker et al., 2007). They found that polycomb group genes Eed and Phc1are both bound by Oct4 and Nanog, are downregulated upon downregulation of Oct4 and Nanog, and all four factors target the promoters of the same set of genes, implicating them as partners in repressing genes associated with development. While gene expression or transcription factor binding studies alone are not sufficient to identify regulatory transcription factor-target interactions, this integrative approach allowed for prediction of novel regulators and regulatory networks controlling ES cell fate, some of which were verified using RNAi combined with gene expression analysis.
To further expand the core transcriptional regulatory network of pluripotency, Kim et al. used a modified ChIP-chip to examine the function of six additional transcription factors (Klf4, c-Myc, Nac1, Zfp281, Dax1 and Rex1) in mouse ES cells (Kim et al., 2008). They showed that regulation of target genes correlates with the extent of promoter occupancy by multiple factors: promoters bound by few factors tend to be repressed, while promoters bound by more than four factors are largely active in the pluripotent state and become repressed upon differentiation. Interestingly, while the majority of the nine factors tested share similar targets, c-Myc and Rex1 occupy promoters of a distinct subset. In addition, c-Myc target genes are associated with the active histone mark H3K4me3 and are more frequently expressed than targets of other factors in ES cells. These results are in agreement with the proposed function of c-Myc in the widespread maintenance of active chromatin (Knoepfler et al., 2006) and suggest a distinct function of c-Myc in regulation of chromatin accessibility in pluripotent cells. In a similar study, Chen et al. used ChIP-seq to determine binding sites of 13 sequence-specific transcription factors (Nanog, Oct4, STAT3, Smad1, Sox2, Zfx, c-Myc, n-Myc, Klf4, Esrrb, Tcfcp2l1, E2F1 and CTCF) and 2 transcriptional regulators (p300 and Suz12; Chen et al., 2008). They found that the majority of genes upregulated in ES cells are bound by either Nanog, Oct4, Sox2, Smad1 and STAT3, or c-Myc and n-Myc. Smad1 and STAT3 share many target sites with Nanog, Oct4 and Sox2, integrating BMP and LIF signaling pathways, respectively, with the core ES cell transcription regulatory network. These two studies demonstrate that gene clustering based on combinatorial transcription factor occupancies has the potential to predict ES-cell specific gene expression.
ChIP-chip, ChIP-PET and/or RNAi in combination with gene expression profiling have proven to be valuable methods to dissect transcriptional regulatory networks. Nevertheless, they all require prior knowledge of the relevant transcription factors. With computational approaches, however, it is possible to identify novel transcription factors using only gene expression profiling data (GuhaThakurta et al., 2002; Liu et al., 2001; Roth et al., 1998; Zhu et al., 2002). Genes with co-regulated expression often share cis-regulatory motifs, corresponding to transcription factor binding sites, in their promoter and enhancer regions (MacIsaac and Fraenkel, 2006; Pennacchio and Rubin, 2001). Using a computational approach, we recently identified DNA motifs that are over-represented in the cis******-acting regions of genes highly expressed in pluripotent cells (Grskovic et al., 2007). These motifs include the binding sites for transcription factors Oct4, Sox2 and Myc, as well as several novel motifs. The analysis of one of the novel motifs identified transcription factor Nfy as a regulator of gene expression in ES cells that is required for their proliferation (Grskovic et al., 2007). Nfy had not previously been implicated in regulation of pluripotency. In addition, gene expression profiling failed to detect Nfy as enriched in pluripotent cells, possibly because one of the Nfy subunits, Nfya, has two splicing isoforms. During ES cell differentiation, the expression of the two isoforms changes in opposite directions (Grskovic et al., 2007), which would be overlooked if expression levels of only common exons were analyzed, as is normally the case with standard microarrays. Therefore, it is important to use unbiased gene profiling approaches to obtain a more comprehensive description of the pluripotent transcriptome.
4. Novel genes and splice variants
A limitation of gene expression studies using standard microarrays is that novel non-annotated genes may not be probed for in the array platform used. A further limitation is that standard microarrays do not generally distinguish between differentially spliced isoforms (see above). Studies of the ES cell transcriptome that did not rely on microarrays revealed that ES cells express numerous uncharacterized or novel transcripts (Araki et al., 2006; Sharov et al., 2003). A large number of ESTs and SAGE tags with no match to a known gene were found to be abundantly and specifically expressed in ES cells (Brandenberger et al., 2004; Richards et al., 2004). In addition, the analysis of gene trap screens revealed thousands of novel exons and genes, many of which are found in gene trapping hotspots associated with loci significantly expressed in ES cells (Roma et al., 2007). Further studies identified naturally occurring antisense transcripts for several pluripotency genes, including Oct4 and Nanog, suggesting that sense-antisense pairing may have a role in regulation of pluripotency (Richards et al., 2006). It should be noted that antisense transcription is not specific for ES cells: recent analysis of global transcription in mouse revealed that a large proportion of the genome produces transcripts from both strands (Katayama et al., 2005).
A next step will be to characterize novel genes expressed in ES cells and evaluate their role in the regulation of pluripotent cells. To this end, Kunarso et al. have analyzed several novel transcripts predominantly expressed in ES cells and found that they represent novel protein-coding genes or intergenic splicing isoforms (Kunarso et al., 2008). Using combined computational and experimental approaches, Pritsker et al. have identified alternative splicing of numerous genes in ES cells, with high levels of alternative splicing in components of signaling pathways that are functional in stem cells (Pritsker et al., 2005). While the identification of novel transcripts and alternative splicing isoforms increases the complexity of the pluripotent transcriptome, future work should reveal what role, if any, they play in maintaining the pluripotent state of ES cells.
5. Is everything transcribed? In every ES cell?
The increase in the number of expressed genes as well as a high number of novel transcripts in ES cells may be due to two possibilities: 1) the transcriptome of ES cells only appears to be more complex than that of other cell types, due to the extensive investigation of ES cells; or 2) ES cells do express a higher number of genes compared to differentiated cells. To investigate these two possibilities, Efroni et al. used tiling microarrays to compare genome-wide transcription profiles of ES cells and neuronal progenitor cells (Efroni et al., 2008). Their analysis revealed genome-wide hypertranscription specifically in ES cells, including expression of normally silent, noncoding regions. In addition, several tissue-specific genes are expressed at low levels in ES cells, suggesting their expression might occur stochastically within the population (Efroni et al., 2008). Increased global transcription in ES cells is in agreement with specific features of the ES cell chromatin, such as a “loose” chromatin structure and the presence of bivalent chromatin marks (Azuara et al., 2006; Bernstein et al., 2006; Meshorer et al., 2006; Niwa, 2007). In addition, ES cells contain higher levels of acetylated H3K9 and methylated H3K4, markers of open chromatin (Azuara et al., 2006). Therefore, it seems that the increase in global transcription in undifferentiated ES cells is an inherent characteristic of ES cells (see Figure 2). However, it is still not clear whether this hypertranscription is simply a consequence of the chromatin properties of ES cells, or whether it has a major role in regulation of pluripotency and self-renewal of ES cells. Furthermore, it is possible that some of the hypertranscription detected in ES cells is derived from heterogeneity in the cell population.
Genome-wide transcriptional profiling typically requires a high number of cells and has therefore been performed using populations of ES cells. However, recent reports have demonstrated cell-to-cell differences in the expression patterns of several genes among undifferentiated ES cells, suggesting that ES cells are not a homogeneous population (Carter et al., 2008; Chambers et al., 2007; Toyooka et al., 2008). Indeed, Toyooka et al. have identified two populations within ES cells that differ in Rex1 expression level and have distinct differentiation potential (Toyooka et al., 2008). Similarly, Nanog is undetectable in a subpopulation of undifferentiated ES cells (Chambers et al., 2007). This subpopulation of cells is predisposed to differentiate but may remain undifferentiated and can subsequently re-express Nanog. These results show that ES cells are a heterogeneous cell population consisting of different types or states of cells. It has been suggested that ES cells may exist in a metastable state which fluctuates between a Nanog-Oct4-Sox2 stabilized state, where differentiation-associated transcription is maintained below threshold levels, and a Nanog-deficient state, where an increase in specific signals, such as phosphorylated-Erk, may trigger differentiation-associated transcriptional networks (Silva and Smith, 2008). Future work should reveal if heterogeneity of undifferentiated cells is limited to the cells tested, or if it is an intrinsic property of all ES cells.
The differential expression of genes in subpopulations of ES cells may be a product of stochastic gene expression, or it may be regulated by an as yet unidentified mechanism (for example, linked to the cell cycle stage). It is also possible that only a subset of undifferentiated ES cells in culture retain full pluripotency. Future work focusing on single-cell transcriptome analysis should clarify the profile and potential of different subpopulations of ES cells.
6. Differences between mouse and human ES cells
Pluripotent mouse and human ES cells by definition share the fundamental properties of self-renewal and the ability to give rise to cells of all three germ layers. Therefore, it is not surprising that they both express components of the core transcriptional network of pluripotency, such as Oct4, Sox2 and Nanog. Nevertheless, there are important differences in the targets of Oct4, Sox2 and Nanog between mouse and human ES cells (Boyer et al., 2005; Loh et al., 2006). These differences are reflected in the divergence between mouse and human transcriptomes, and correlate with differential cytokine and growth factor requirements, growth rates and marker expression between the ES cells of the two species (Wei et al., 2005). For example, human ES cells lack transcripts of the LIF signaling pathway, while this signaling pathway is expressed, active and required for mouse ES cells. Similarly, several components of the FGF pathway are expressed in human ES cells while being absent from mouse ES cells. Cell surface markers SSEA1, SSEA3 and SSEA4 are also differently expressed in mouse and human ES cells (Thomson et al., 1998).
The differences between mouse and human ES cells may represent species-specific transcriptional programs. Alternatively, mouse and human ES cells may represent different stages of early development. Consistent with the latter hypothesis, pluripotent epiblast cells (EpiSCs) can be derived from post-implantation mouse embryos (Brons et al., 2007; Tesar et al., 2007). Unlike mouse ES cells, but similarly to human ES cells, mouse EpiSCs require Activin/Nodal and FGF signaling for their derivation and maintenance (reviewed in (Yu and Thomson, 2008)), and are able to differentiate into trophectoderm. In addition, there is a greater overlap between genes bound by Oct4 in human ES cells and mouse EpiSCs than between human and mouse ES cells (Tesar et al., 2007). Therefore, it is possible that mouse and human ES cells correspond to different stages of embryonic development. Future studies of EpiSCs may reveal the extent of species- and stage-specific differences between the mouse and human pluripotent transcriptome.
In summary, there is substantial evidence that both conserved and divergent pathways act to regulate self-renewal and pluripotency in mouse and human ES cells. Even though mouse ES cells are an excellent model to study developmental processes and diseases, care must be taken when extrapolating results obtained using mouse ES cells to human ES cells.
7. Expression of the pluripotent transcriptome in cancer
Cancer cells are capable of self-renewal and possess high phenotypic plasticity, which are the major features of stem cells. In fact, it is thought that some cancers may arise from aberrant stem or progenitor cells, or that cancer cells develop by progressive de-differentiation of their normal counterparts. Such de-differentiation might occur if a particular combination of stem cell-associated factors is expressed in cancer cells, perhaps in a manner analogous to the generation of iPS cells. Indeed, ectopic expression of Oct4 inhibits progenitor cell differentiation and causes dysplasia in epithelial tissues (Hochedlinger et al., 2005). It should be noted, however, that somatic cancers have not been conclusively shown to express Oct4: the expression reported by several groups (Atlasi et al., 2007; Ezeh et al., 2005; Gibbs et al., 2005; Iki and Pour, 2006; Ponti et al., 2005; Tai et al., 2005) was recently attributed to the expression of an Oct4 pseudogene or of Oct4B, an isoform of Oct4 that is not associated with pluripotency (Cantz et al., 2008). Recently, two groups have analyzed the expression of particular ES cell-associated gene sets in various tumors (Ben-Porath et al., 2008; Wong et al., 2008). Interestingly, they found that the ES cell-like transcriptional program is active in several human tumors and is correlated with metastasis and mortality. This ES cell-like transcriptional program includes genes with roles in cell cycle, signaling, transcription, DNA repair, stress response and differentiation. The targets of Oct4/Sox2/Nanog are found to be expressed in some cancers despite the lack of Oct4 and Nanog, indicating that the pluripotency-associated transcriptional program may be activated by alternative mechanisms. Furthermore, Wong et al. have shown that c-Myc can activate the ES cell-like transcriptional program in adult epithelia and in an in vivo model of human epithelial cancer, leading to an increase in the number of tumor-initiating cells. In particular, expression of c-Myc led to induction of Sox2 and repression of differentiation regulators, such as Hox genes. These findings are in agreement with the role of c-Myc as one of the key transcription factors involved in inducing pluripotency (Lowry et al., 2008; Maherali et al., 2007; Meissner et al., 2007; Park et al., 2008; Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Wernig et al., 2007; Yu et al., 2007) and may relate to the function of c-Myc in the widespread maintenance of active chromatin (Knoepfler et al., 2006). It should be noted, however, that pluripotent cells can be derived from somatic cells in the absence of c-Myc, albeit with a lower efficiency (Nakagawa et al., 2008; Wernig et al., 2008).
Some germ cell tumors, which are thought to arise from transformation of PGCs (Stevens, 1967), have transcriptional profiles similar to ES cells, including the expression of Oct4, Sox2 and Nanog (Santagata et al., 2007; Skotheim et al., 2005; Sperger et al., 2003). It is important to note that the expression of Oct4, Sox2 and Nanog is not sufficient to cause tumors: normal PGCs express all three factors at levels similar to those in ES cells. To identify genes with possible role in germ cell tumor formation, we compared the expression profiles of PGCs to pluripotent stem cell types (ES cells, EG cells and ICM) and to publicly available expression data from germ cell tumors (Wei et al., submitted). We found that the genes absent from PGCs but highly expressed in pluripotent cell types are up-regulated in some germ cell tumors. Among these is Klf4, a transcription factor that functions with c-Myc to induce pluripotency in somatic cells. It is possible that Klf4, similarly to c-Myc, activates the ES cell-like transcriptional program and induces tumor-initiating cells. In agreement with this model, expression of Klf4 in basal keratinocytes leads to squamous epithelial dysplasia (Foster et al., 2005). These results suggest that similar mechanisms may govern the conversion of PGCs to the pluripotent stem cell state (EGs) and germ cell tumorigenesis. Further functional studies should address the role of regulators of pluripotency in tumorigenesis. Detailed characterization of the ES cell-like transcriptional networks in cancer cells may reveal novel diagnostic markers and potential therapeutic targets (see Figure 3).
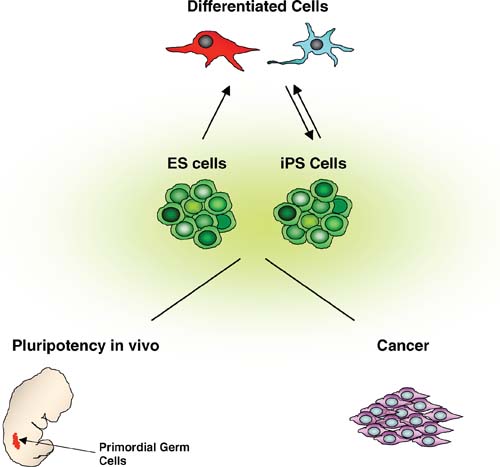
The development of high-throughput genome-wide expression analyses has led to the identification of the ES cell transcriptome. There are still several issues that need to be addressed, such as permissive transcription, stochastic gene expression and heterogeneity in the population of undifferentiated ES cells (see text). Insights gained by analyzing the transcriptome of pluripotent cells will feed into, and feed back from, research focusing on: a) the role of pluripotency in vivo, in particular through the study of the function of regulators of pluripotency in PGCs, b) the role of ES cell-like gene expression in cancer, which may lead to development of novel diagnostic and prognostic markers as well as new therapies; and c) the regulation of ES cell self-renewal and differentiation and of somatic cell reprogramming to the pluripotent state.
8. In vivo transcriptomics – a germline connection?
The majority of the transcriptional profiling studies aimed at understanding pluripotency were performed using pluripotent stem cells grown in vitro, mostly because this type of analysis requires large numbers of cells that are difficult to obtain in vivo. However, understanding the transcriptome of pluripotent cells in vivo may provide novel insights into the significance and regulation of pluripotency. Recently, we performed a comparative study of the gene expression profiles of mouse pluripotent stem cells and the cells in the embryo from which they are derived (Wei et al, submitted). We found that PGCs express the global transcriptional program for pluripotency at levels similar to ES cells. Furthermore, PGCs express high levels of pluripotency regulators such as Oct4, Sox2, Nanog, Sall4, N-Myc, Utf1 and Rex1, indicating that some of the factors and mechanisms that maintain pluripotency in early embryos and ES cells may also be operating in the embryonic germline.
The evolution of the molecular mechanisms that regulate pluripotency has not been studied in detail. The sequence and function of known regulators of pluripotency, such as Oct4 and Nanog, do not appear to be well conserved outside of mammals. Putative orthologs of Oct4 or Nanog have been identified in Xenopus and chicken (Lavial et al., 2007; Morrison and Brickman, 2006), but they are more broadly expressed than in the mouse embryo. XenopusOct4 is required for maintaining the numbers of uncommitted cells during gastrulation, suggesting an ancestral role for Oct4-like proteins in preserving the undifferentiated state in early embryos (Morrison and Brickman, 2006). Oct4 and Nanog promote self-renewal in chicken ES cells, but their function in the chick embryo has yet to be determined (Lavial et al., 2007). It is interesting to note that chicken Oct4 and Nanog are expressed in PGCs, and that the axolotl also contains an Oct4 gene expressed in gastrulating embryos and oocytes (Bachvarova et al., 2004). It is possible that the program for pluripotency evolved in mammals, from molecular components already present in low vertebrates, to inhibit somatic differentiation and preserve germline competence until the germline is induced and fully developed. If that is the case, regulators of pluripotency are expected to have essential roles in PGC development. Interestingly, Nanog-deficient PGCs fail to mature into germ cells (Chambers et al., 2007). As the timing of the defect corresponds to the timing when PGCs undergo extensive epigenetic changes (imprint erasure, genome-wide demethylation and X chromosome inactivation in female cells), it was suggested that Nanog functions by maintaining PGCs in a state of minimal epigenetic control, similar to the ES cell state (Chambers et al., 2007). In addition, inactivation of Oct4 specifically in PGCs leads to a loss of germ cells (Kehler et al., 2004). Further studies of gene expression and dissection of the transcriptional regulatory networks in PGCs should shed light on the role of pluripotency in the germline (see Figure 3).
9. Concluding remarks
It is becoming apparent that a detailed understanding of regulation of gene expression in pluripotent cells will continue to make important contributions to developmental, stem cell and cancer biology. The emerging picture is one where specific transcription factors, operating through self-enforcing mechanisms, maintain the pluripotent state in the backdrop of a loose, non-restricted chromatin. Increased global transcription in ES cells may be a by-product of loose chromatin. Alternatively, increased transcription may be required to maintain the chromatin in accessible, pluripotent state. Fluctuations in the transcriptional regulatory network may cause ES cells to oscillate between different states with distinct propensities for self-renewal or differentiation. It will be of interest to investigate whether probabilistic behavior is central to ES cell pluripotency, and if so how the probabilities of self-renewal or differentiation can be modulated (Ramalho-Santos, 2004).
Accessible chromatin and fluctuating transcriptional networks could allow for stochastic gene expression that escapes deterministic transcriptional regulation. Therefore, it will be necessary to annotate and characterize novel transcripts found in ES cells and determine whether they have a role in regulating pluripotency. The development of new, more sensitive methods, such as deep sequencing, should allow for unbiased identification of low-abundance transcripts, as well as newly emerging small RNA species. As even more sensitive large-scale analytical techniques to assess gene expression are developed, it will be important to analyze gene expression in single ES cells, and ideally to correlate individual transcriptomes to developmental potential. The results of transcriptome analysis will have to be complemented by proteome and RNAome analysis to account for the role that post-transcriptional regulation may play in pluripotent cells. Several miRNAs were found to be predominantly expressed in ES cells, and their analysis revealed that miRNAs play crucial roles in ES cell self-renewal and differentiation (reviewed in (Blakaj and Lin, 2008). Future work will determine the role that miRNAs may have on modulating the transcriptional regulatory network of pluripotency (Marson et al., 2008). Finally, it will be crucial to investigate the role(s) of pluripotency in vivo in the germline and in tumor development.
Elucidation of mechanisms governing pluripotency should deepen our fundamental understanding of early embryogenesis and germline development. It will also inform our efforts to generate disease- or patient-specific pluripotent stem cells and to target differentiation towards cell types of therapeutic interest (see Figure 3). Finally, insights gained into the regulation of pluripotency may facilitate the identification of new cancer markers and therapeutic targets. Clearly, the pluripotent transcriptome is “poised” for significant impact in a variety of fields in the years ahead.
Acknowledgements
We would like to thank Robert Blelloch, Kevin Corbit, Erica Jackson, Collin Melton, Yuki Ohi, Jeremy Reiter and Yangming Wang for helpful comments on the manuscript. M.G. was supported by CIRM, and M.R.S. by CIRM, JDRF and IRM.

